Projects
Contents
- Project Outline
- Idea 1. Bio-Data Science in Python
- Idea 2. Bio-Data Science in Julia
- Idea 3. Web-development via RStudio’s blogdown
- Idea 4. Interactive visualization for a biological dataset
- Idea 5. Deep learning in biology.
- Idea 6. Develop a new tool for a problem arising in biology
- Idea 7. Dashboards for your research project
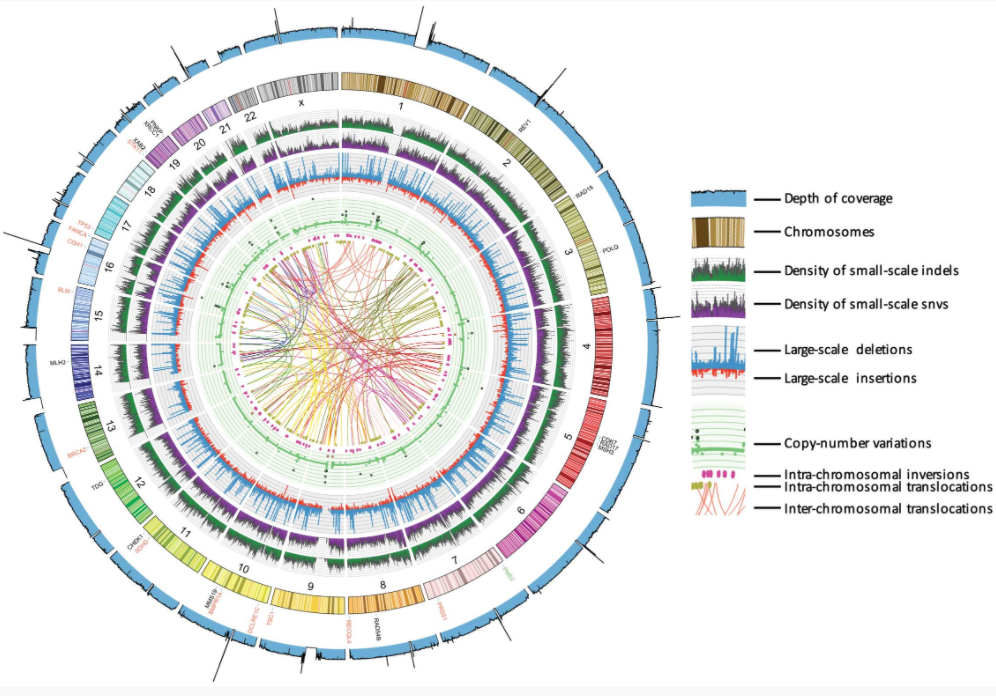 Circos plot depicting the patterns of genetic variations for a patient with Li-Fraumeni syndrome Li et al. (2015)
Circos plot depicting the patterns of genetic variations for a patient with Li-Fraumeni syndrome Li et al. (2015)
[Graduate (610) & Genomics Diploma (530) Students]
There is a small project that is worth \(10\%\) of the overall grade. It
should be the equalivalent work as an assignment.
Project Outline
You can work in groups, but the larger the group, the more that is expected.For those team members that go above and beyond, remember the \(+1\) mechanism!
By mid-semester (date), you or your group should submit to the instructor by Slack a one page outline of your proposal.Pick a topic close to your research heart. Perhaps the resulting tool could actually be useful to you and your research down the road.
The projects can be submitted at the end of term; they will be displayed for prosperity on on this website.With students’ permissions of course.
A mixture of media can be used including text documents (eg Google Docs), images (eg Google Drawings), R Markdown, or R Notebooks. Code should be able to be executed by the instructor, and the results should be as reproducible as possible.
A small manual or guide to your project should be submitted.Have fun with it!
The following are just suggestions and you are not restricted to these topics.
Idea 1. Bio-Data Science in Python
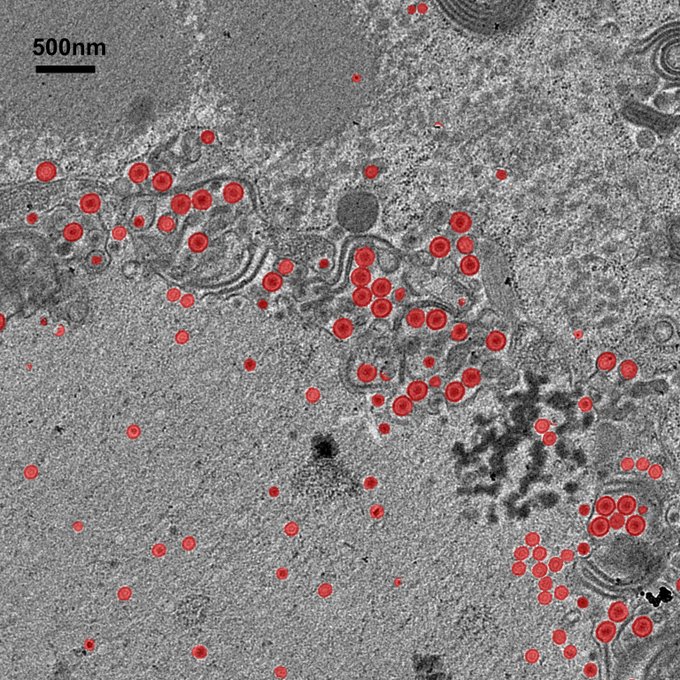 Automated identification of viruses in EM images Ariadne AI Inc.
Automated identification of viruses in EM images Ariadne AI Inc.
Many people ask why \({\tt R}\) and not \({\tt Python}\), a programming language often used in bioinformatics and elsewhere. There is no simple answer to this. Both have their strengths and a good Biodata Scientist will learn both. The goal of the project could be to learn the basics of \({\tt Python}\) You would be expected to show proficiency in the language to approximately the same level in the course for \({\tt R}\). You can be creative and choose to visualize complicated biological data or, for example, re-do questions from the assignment in \({\tt Python}\) rather than \({\tt R}\).
Idea 2. Bio-Data Science in Julia
Julia is relatively new language that is increasingly used in bioinformatics. Many people think this is going to be a very important and ubquitious language. The goal of the project could be to learn the basics of \({\tt Julia}\). You would be expected to show proficiency in the language to approximately the same level in the course for \({\tt R}\). You can be creative and choose to visualize complicated biological data or, for example, re-do questions from the assignment in \({\tt Julia}\) rather than \({\tt R}\).
Idea 3. Web-development via RStudio’s blogdown
This entire course was developed in RStudio’s blogdown. This includes the course website and the course lecture slides. blogdown uses the R markdown language for making fancy text. The goal of this project could be to develop using this system a personal academic web page for yourself or your lab.
Idea 4. Interactive visualization for a biological dataset
The goal of the project is to explore visualization in R using Shiny, produced by RStudio. Shiny allows users to quickly produce and publish interactive visualizations. Here the project might involve developing a Shiny app for a particular dataset that you are interested inI can also make recommendations for publicly accesible datasets.
. Although you are welcome to develop a Shiny tool from scratch, it might be easier to start wth an example from the Shiny Gallery.
Idea 5. Deep learning in biology.
Develop a deep neural network for a problem related to biology. This can be done using one of several packages for deep learning in \({\tt R}\). For example, you can use KERAS in RThere are other options besides KERAS. So if you have an interest in a different deep learning package, go for it!
. There are nice tutorials online. Maybe … maybe… I will allow you to use my lab’s GPU cluster but start with your RStudio Cloud. I can help you find an appropriate biological dataset and/or assist you with defining a good learning question.
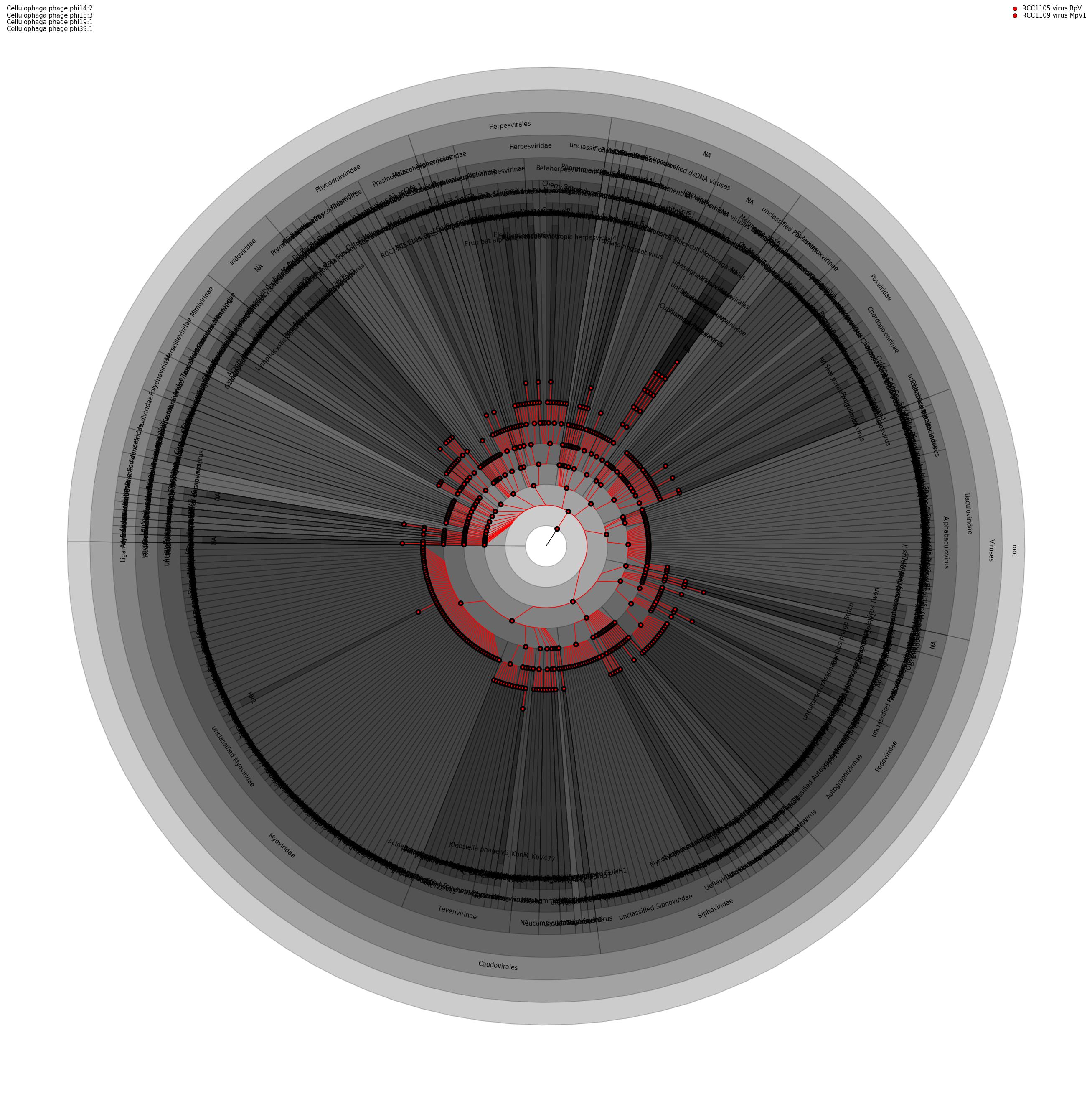 Visualizaton of the Barbadian reef microbiome. From Shawn Simpson in the Hallett Group
Visualizaton of the Barbadian reef microbiome. From Shawn Simpson in the Hallett Group
Idea 6. Develop a new tool for a problem arising in biology
Perhaps you already have some computational challenges w.r.t. your own research project. You are welcome to use this as the basis of your project. Here you would just define one such problem and show how tools from bioinformatics, computational biology or data science can be used to address the problem. Microscopy has many interesting challenges and I can help you get started.
Idea 7. Dashboards for your research project
Dashboards are really nice tools for bringing together different interactive visualizations and data analyses. A very popular one currently in John Hopkin’s Coronavirus Resource Centre. These are often built with Shiny.