Assignment 2
Please note that due dates can be found in the Syllabus; submission instructions can be found on the Assignment Instructions page. In this assignment, you can submit a Google Doc (or other text editor, pictures, etc.) but also your \({\tt R}\) code via Google Drive. Aki will go over the submission process in the lab.
You might consider (but it is not mandatory) using R Markdown to write your answers.
\({\bf 67}\) total marks.
Question 1 [10 points] A. Re-express the following small program using pipes.
c <- 18
f <- c*(9/5) + 32B.
x <- 1e10
y <- sqrt( factorial(x) )C.
x<-3; mu<-5; sigma <-7
nrm <- ( 1 / (sigma * sqrt( 2 * pi ))) * exp( -(1/2) * ( (x - mu)/ sigma )^2)Hint:
p <- c( first = "a", second = "b", third = "c")D. What is this equation?
E.
site <- 15
gsub(";", "/", paste(paste( paste( "~hallett", paste("raw", "tara", sep = ";"), sep = ";"),
paste("site", site, sep="_"), sep=";"),
"dna_seq.fa", sep=";"))
## [1] "~hallett/raw/tara/site_15/dna_seq.fa"Question 2 [5 points]
In total, how many black, african or asian women are included in the \({\tt small\_brca}\) dataset?
Create a histogram of all asian individuals grouped by age. Each bar in the histogram should have two colours corresponding to male and females. (For example, red might be used for the fraction of women in some age group while blue is used for men in that same age group). Choose the number of bins (or binsize) in a way that makes your plot informative.
Show code for both parts.
Question 3 [7 points]
How many participants are there? (Careful: not samples but participants.) Show code.
List all of the \({\tt participant}\) that have more than one sample. Show code.
For the participants identfied in 4b, create a scatter plot with the tumor on the \(x\) axis and the normal on the \(y\) axis (as specified by the \({\tt tumor}\) variable in the tibble) with points representing the expression of gene \({\tt ESR1}\).
Show your code for each step.
Question 4 [23 points]
Show your code for each step below.
Part a [1 point] Load the \({\tt tidyverse}\) library and load the \({\tt small\_brca}\) dataset.
Part b [1 point] Write code that determines the number of rows (observations) and columns (variables) in \({\tt small\_brca}\).
Part c [1 point] Write code that displays the column names (names of variable) for columns \(21\) to columns \(24\) inclusive.
Part d [2 points] What is the mean expression level of FOXC1?
Part e [3 points] How many samples are there with FOXC1 expression levels above \(245\)?
Part f [5 points] Create a new tibble called \({\tt bw}\) that contains only those rows (observations) from \({\tt small\_brca}\) that correspond to tumor samples of BLACK OR AFRICAN AMERICAN women.
Part g [5 points] Using \({\tt small\_brca}\), select the variables \({\tt participant}\), \({\tt vital\_status}\), and all genes except \({\tt RRM2}\).
Part h [5 points] Create a new variable called \({\tt triplenegative}\) in the tibble \({\tt small\_brca}\) that is TRUE (logical) if the sample is “Positive” for \({\tt er\_status\_by\_ihc}\), and not “Positive” for \({\tt her2\_fish\_status}\). Otherwise, in all other cases it is FALSE.
Question 5 [12 points]
Using the \({\tt small\_brca}\) tibble, create the following plot.
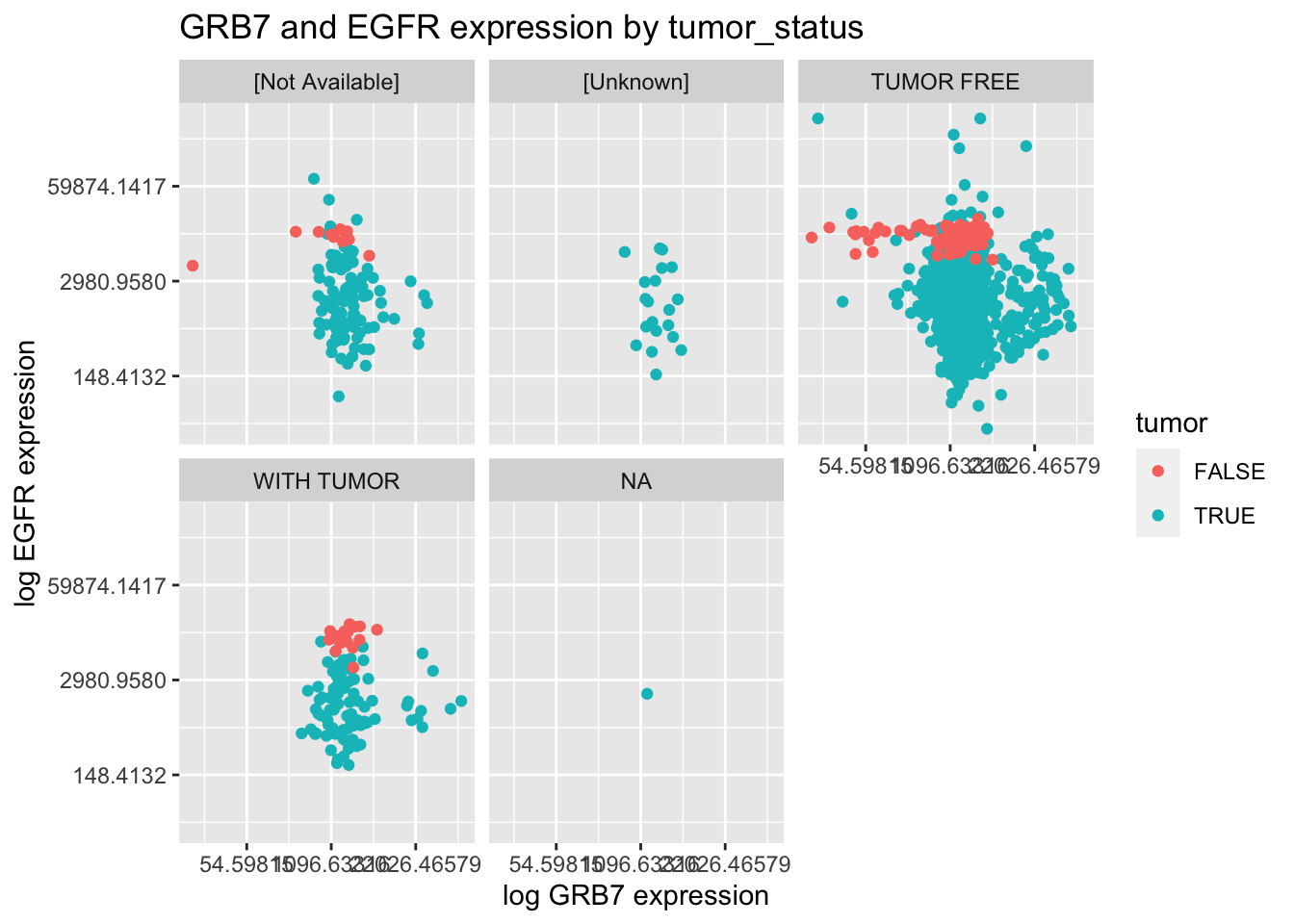
Question 6 [5 points]
The variable \({\tt race}\) in \({\tt small\_brca}\) has several different possible values including “WHITE”, “ASIAN”, “BLACK OR AFRICAN AMERICAN”, “AMERICAN INDIAN OR ALASKA NATIVE”, “[Not Available]”, “[Not Evaluated]”, or NA.
It is better to simply use NA instead of“[Not Available]”, “[Not Evaluated]”, or NA. It will simplify your code in downstream analyses.
Write R code to create a new \({\tt race\_modified}\) variable where all samples with “[Not Available]”, “[Not Evaluated]” are changed to NA (Note: you shouldn’t use “NA”, but NA. They are not the same)
Question 7 [5 points] Pick one of the \(50\) genes from \({\tt small\_brca}\) uniformly randomly (see the \({\tt runif}\) function). Using the NCBI, provide the following information or note that it is not availalbe:
10a Full name of the gene and the official name according to the HGNC.
10b First time it was reported in the literature.
10c Where it is located in the genome.
10d Its \({\tt gi}\) acccession code or codes (if it has been modified).
10e The number of exons.
10f The number of alternative transcripts that have been record.
10g Its full amino acid sequence. Provide its protein ID.
10h Its protein structure, if known.
Good luck!