Overview
Course objectives
Our fundamental goal is to ensure you have the necessary bioinformatics skills, both in terms of tools and underlying analytic approaches,in order to fully participate in modern life science approaches.
Biology is becoming increasingly quantitative.
Instructor
Mike Hallett
TBA
Office Hours TBA
hallett.mike.t@gmail.com
@hallettmichael
Course
TBA
TBA2022
TBA
Zoom
Slack
Github repo
Lectures
RStudio Cloud
Labs
TBA
TBA
TBA
Zoom
Office Hourse: TBA
There are many reasons for this including the development of high-throughput profiling approaches. Nucleotide sequencing, protein/lipid/small molecule mass spectrometry, and cellular/subcellular microscopy, but the list of
new technologies continues to expand rapidly.
All areas of life science research from basic biolgy to human health-related efforts have been fundamentally changed by this influx of technology.
[Data Science] A single -omics experiment now can produce a staggering amount of data which would be impossible to sift through using only human hands and eyes. The ability to develop software and analytic approaches to wrangle with this data, clean it up, kick it into shape and visualize it in a way that is both informative and honest is super important skill set to have as a modern life science student. This course will give you the fundamentals of data science in the context of many examples including metagenomics of ocean ecologies and transcriptomics of breast tumors.
[Bioinformatics] The ability to profile complete genomes, microbiomes, proteomes, interactomes and other “-omes” creates a need for computational infrastucture to capture and organize this data. This is often referred to as bioinformatics, and is focused on the development of tools, portals and databases to make this information available to all life scientists. This course will give many examples primarily centered on the resource available at the NCBI.
[Computational Biology] Computational biology concerns itself with the development of new analytic techniques, typically expressed as computer programs, to explore data. For example, if you were generating many high-content microscopy images and were interested identifying specific events (eg expression of a rare cellular or subcellular phenotype), you would benefit from developing a machine learning algorithm to sift through the gigabytes of images automatically. At the end of this course, you wil understand the basics of machine learning including (Hidden) Markov Models, linear and logistic regression, and deep convolution neural networks.
Course materials
 Free on-line textbook Grolemund Wickham
Free on-line textbook Grolemund Wickham
[Text Book] Throughout much of the course, we will follow
- Hadley Wickham and Grolemund, R for Data Science (NZ: O’Reilly Publishers Inc., 2017), https://r4ds.had.co.nz/. (free) online and from Amazon]
Although th electures are styled following this R based textbook, we recommend
if you plan to complete the course primarily using Python. This will give you tips for that language.
Similarily, if you would like to complete the course using the Julia language, we recommend
as a starting point. In general, the literature for Julia is sparser than for R and Python. We really recommend Julia only for those with significant prior programming experience.
However note that all of these are general data science book and not specific to biology and the life sciences. For several lectures, I will provide additional reading to cover the biology here.
 Not-free Python textbook McKinney
Not-free Python textbook McKinney
[Software] The course will heavily utilize several software packages
Google Gmail (free) For security reasons and for simplicity, we ask each student to obtain a gmail account for the course. You will need to send it to the bioinfo.western@gmail.com at the beginning of the course.
Google Drive (free) We use the cloud-based text editor, drawing tool, spreadsheets, and storage offered by Drive to create and share files.
YouTube (free) All lectures are online at our YouTube channel.
 Free Julia documentation Julia
Free Julia documentation Julia
Slack (free)When you send me your gmail account, you will be added.
Zoom (free) All lecture slots and TA sessions are on our Zoom channel available at the link above. When you have given us your gmail account, we will add you to Slack and you will see the password for the Zoom channel.
RStudio Cloud (~$28.00) This is a beautiful Interactive Development Environment (IDE) for the language . It is based in the cloud, so you will not have to install any software. All of the lecture notes, code and datasets will be available to you at the website.
There are other ad hoc tools that we will use throughout the course, but they are all free.
Total cost for the course is ~$28.
[RStudio for Python too] Whether you plan to complete the course using R or Python (or Julia), we still recommend using RStudio. You will learn about Jupyter, a web based IDE which was designed for Julia, Python and R, but RStudio still has some advantages, especially with packages like reticulate which we will learn about.
[Alternatives to R] If students would prefer to complete the course using Python or Julia, they may do so. Note however that the lectures will still be given in R, and some coding exercises may require that you program in R. There are several reasons. For example, we make data available to you in an R datastructure, or we use a package that is specific to R which does not have an analog in the other language. Most packages do have versions in at least two of the languages. For example, the Keras package for R is actually built on top of the Python pacakge. You may experience increased difficulty installing these packages, compared to our RStudio Cloud version.
[Ease of learning] I would like to stress that R/RStudio is a beautiful and very powerful system. In my opinion, if you are student from the life sciences you will most likely need data science skills the most (over bioinformatics and computational biology). In my opinion, the best tool for you is R. Your needs and goals are not the same as those of acomputer scientist
[Data science vs…] The datascience toolkit for R remains more advanced than Python or Julia. Conversely, if you want to become a computational biologist and develop tools (software based on statitical and computational paradigm), a language like Python is more convenient to program in. If the distinction between data science and computational biology is not clear at this point for you, I would recommend starting with R and perhaps exploring the other languages later.
[You’ll be a bit on your own] I would recommend using Python or Julia only if you are an advanced programmer, or if you already have a significant base in R (by this I mean you are experienced with writing functions, the tidyverse, S4 objects etc.). Note that the TA will have little knowledge outside of R and they do not have the time to solve problems and answer questions. I will try my best but solving conflicts in someone’s conda environment can take hours, so you cannot expect that.
[Pythonic] For people choosing Python, you should use version 3. We recommend that you install anaconda3/conda environments. We also recommend installing and becoming familiar with Jupyter. There are many Interactive Development Environments (IDEs) for Jupyter. I personaly prefer RStudio as it allows R and Python to run together; this will also address some of the aniticipated issues above. If you experience significant difficulties preparing this environment, we ask you to seriously reconsider your choice to use Python.
If you are a relatively advanced programmmer, we recommend the following manual. Otherwise, there are many excellent beginner guides from publishers such as O’Reilly.
[Julia is beautiful.] For people choosing Julia, I personnally use JuliaPro as my IDE (but find myself often using RStudio in the end). We also recommend installing and becoming familiar with Jupyter. I am still a novice Julia programmer and do not have a good overview of the best tools and practices with this language yet. Bonus points if you can lead me to the light here! I do not know if the Julia has the full range of libraries to do all components of this course.
[Hardware] All of the tools listed above are in the cloud. This means that you need minimal computing equipment. It will likely be very hard to program with only a tablet however. Note that you can purchase a Google Chrome notebook for under $300 on Amazon.
Evaluation
Up to 5 pts can be added to your score (out of 100) from bonus awarded to you by students, the instructor or TA. It goes like this: if someone
helps you very significantly with an assignment question or project,
then you inform the instructor
why that person deserves , in a paragraph. Little puzzles in lectures and labs can also earn people . Most s are earned however through answering each other’s questions in the Slack channel for the course.
You can find descriptions and instructions for each exercise on the assignments page.
[Grading Scheme Undergraduates]
| Assignment | Points |
|---|---|
| Quizzes, Puzzles, Opinions (3 x 5 pts) | 15 |
| Midterm (open book 1 hour) | 20 |
| Final Exam (open book 2 hours) | 25 |
| Homework assignments (4 x 10) | 40 |
Total pts: 100
[Grading Scheme Genomics Diploma and Gradaute Students]
| Assignment | Points |
|---|---|
| Quizzes, Puzzles, Opinions (3 x 5 pts) | 15 |
| Midterm (open book, 1 hour) | 15 |
| Final Exam (open book, 2 hours) | 20 |
| Homework assignments (4 x 10) | 40 |
| Project | 10 |
Total pts: 100
A day in the life …
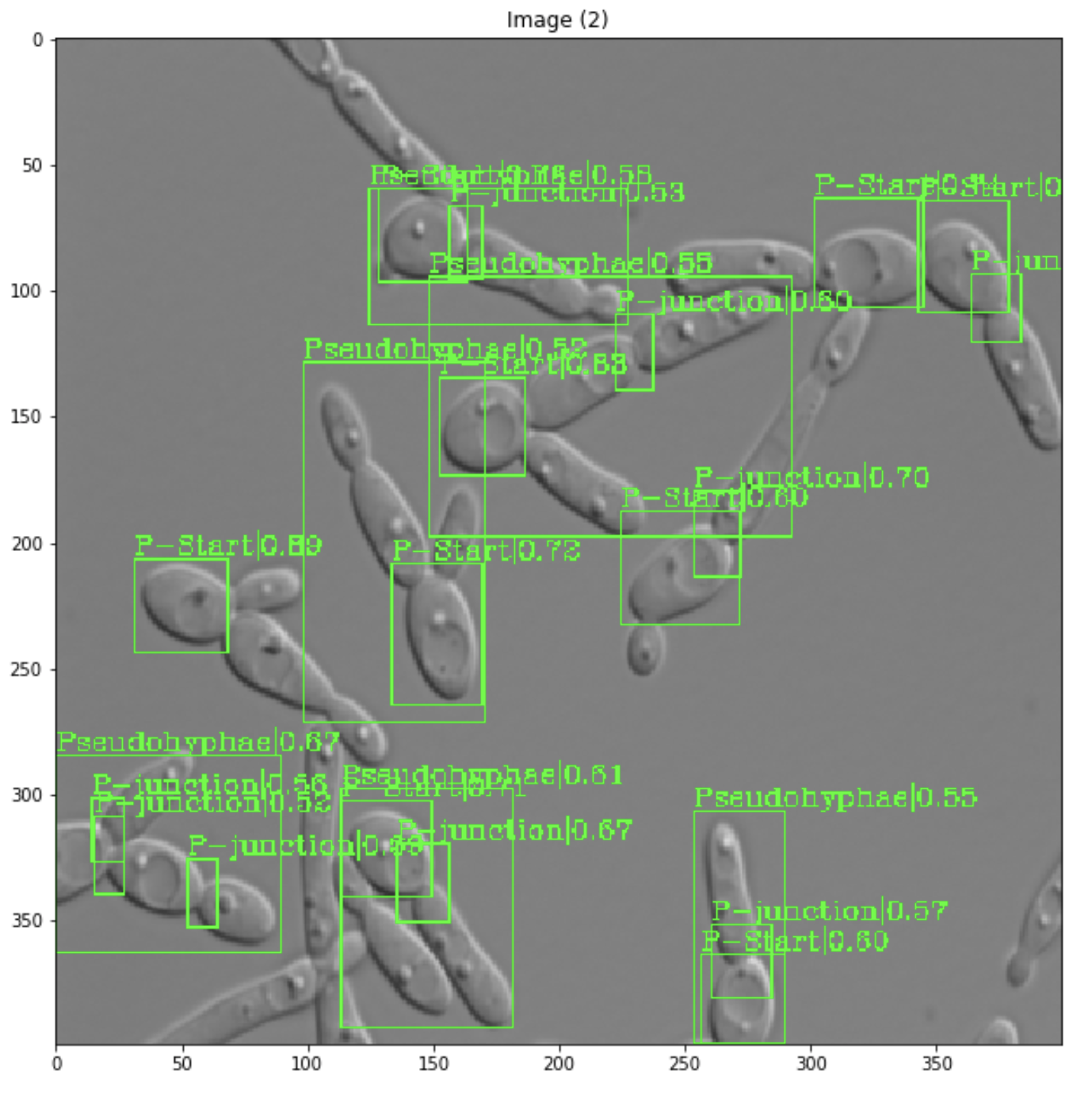 Deep learning-based tool to identify Candida albicans morphologies V Bettauer from Hallett lab
Deep learning-based tool to identify Candida albicans morphologies V Bettauer from Hallett lab
Although covid-19 is still a significant problem, the intention currently is to offer this course in class next term. As I am sure we all appreciate, this could change. The structure of the course is described below.
The video for each lecture is available on our YouTube channel. You should watch the video before the actual lecture.
We have a Slack workspace and you can ask questions in the #biol480 channel. You can get access to Slack by sending your gmail account here: bioinfo.western. Slack is great for asking questions.
The lectures will be used to discuss the material you have read previously (see 1 above). Also, sometimes (8 in total) there will be small quizzess, or I will give a small puzzle, or ask for your opinion on a subject relevant to the material covered to date. These will last ~10 minutes. Otherwise, I will answer questions and provide more depth on specific aspects of the slides. The lectures will be in class.
The labs will be in person but you should bring your computer (if possible). The TA Samira Massahi will answer questions and assist you with the assignments.
Office hours will be in person. I am actually accessible almost always via Slack and find this a more efficient way to communicate. Slack does have video conferencing too that is very easy to use, and is more suitable for one on one discussion.
Submission of assignments, projects and other material will be discussed as we progress through the course.
Time management
As a rule of thumb per week (2 lectures/week),
- readings requires hours;
- lecture on average hours;
- lab hour;
- assignment or project work hours;
- Slack questions or office hours (TA or instructor) hour;
- studying for midterms or exams hour per week.
In total over weeks, this consists (credits at hours each).
Extraordinary circumstances
In the event of extraordinary circumstances and pursuant to the Academic Regulations the University may modify the delivery, content, structure, forum, location and/or evaluation scheme. In the event of such extraordinary circumstances, students will be informed of the changes.
Behaviour
All individuals participating in courses are expected to be professional and constructive throughout the course, including in their communications.
Students are subject to the Code of Rights and Responsibilities which applies both when students are physically and virtually engaged in any University activity, including classes, seminars, meetings, etc. Students engaged in University activities must respect this Code when engaging with any members of the Western community, including faculty, staff, and students, whether such interactions are verbal or in writing, face to face or online/virtual. Failing to comply with the Code may result in charges and sanctions, as outlined in the Code.
Course policies
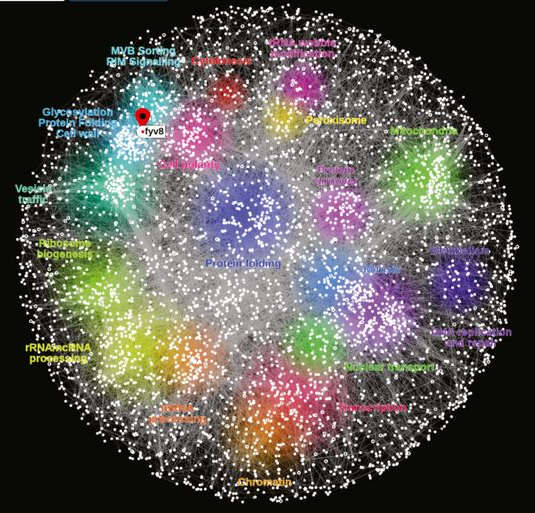 Visualization of the Global Yeast Genetic Interaction Network Usaj et al.
Visualization of the Global Yeast Genetic Interaction Network Usaj et al.
Be nice and honest.. This New York Times article seems appropriate in the covid-19 era: If everyone else is doing it …. You might check out the ethos statement my lab designed; it clarifies everyone’s roles and responsibilities.
In some exercises, we state explicitly that group work is encouraged. In all cases, you must cite whomever helped you and state precisely what they helped you with. If this is not done, this constitutes academic dishonesty. Generally however discussion and interaction is encouraged in the course.
It is challenging to interact via Zoom and Slack. Although not a rule, I prefer if your cameras are on so that we can speak face to face. If that is problematic (eg slow internet, bad hair day), turning the camera on when asking questions is a nice gesture.
Non-Western students and auditors
I am totally happy to allow you to audit or follow the course either in real time, or via the residual online resources. If you want to participate in virtual lectures, labs, office hours, you would need to have our Zoom access code, access to our Slack workspace and Google Drive resources. Please drop me an email.
Action items
Once you have read this entire overview, please email us at the course gmail bioinfo.western with your gmail account, last name, first name and student ID. Make sure it is easy for us to associate the name of your gmail account with your real name (if the username of your gmail account is e.g. “kingofeverything”, it might be hard for us to guess who this is, unfortunately. For security reasons and for organizational simplicity for TA, we would ask that you send a Google mail gmail address.